A pair of Penn State News articles have recently featured our collaboration with Dr. Frank Pugh’s lab to build an automated pipeline for processing ChIP-exo data.
As described in a recent Huck Institutes News article, we have been working with the Pugh lab to build a system for processing and analyzing ChIP-exo sequencing datasets. This system is based on Galaxy, an open, web-based platform for enabling reproducible research that is developed by Dr. Anton Nekrutenko’s lab here at Penn State. Our ChIP-exo Gal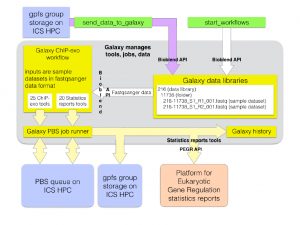 axy “flavor” provides a standardized data analysis pipeline for all ChIP-exo datasets produced by the Pugh lab. Datasets are brought through a complete set of analyses, including quality control, genome alignment, protein-DNA binding event calling, motif-finding, and visualization. The pipeline is also completely automated; once a sequencing run finishes on the Pugh lab’s Illumina NextSeq 500 DNA sequencer, data is automatically transferred to our Galaxy pipeline, which in turn initiates data processing jobs on our ACI-ICS cluster resources. The Galaxy pipeline sends all results and metadata to a custom database and web interface called PEGR (Platform for Eukaryotic Gene Regulation).
axy “flavor” provides a standardized data analysis pipeline for all ChIP-exo datasets produced by the Pugh lab. Datasets are brought through a complete set of analyses, including quality control, genome alignment, protein-DNA binding event calling, motif-finding, and visualization. The pipeline is also completely automated; once a sequencing run finishes on the Pugh lab’s Illumina NextSeq 500 DNA sequencer, data is automatically transferred to our Galaxy pipeline, which in turn initiates data processing jobs on our ACI-ICS cluster resources. The Galaxy pipeline sends all results and metadata to a custom database and web interface called PEGR (Platform for Eukaryotic Gene Regulation).
An earlier article in Penn State News described the network infrastructure that enables our processing systems. We are early adopters of the Penn State Research Network, a high-speed, secure network on campus. This infrastructure enables fast transfer the large datasets produced by the sequencer machine, thereby allowing speedy processing of experimental data.
While this work has been a group effort by both labs, the key people involved in designing & building the framework are Greg Von Kuster (ACI-ICS consultant), Gretta Armstrong, Danying Shao, Bongsoo Park, Will Lai, and Naomi Yamada.



