Our group manuscript, entitled “An integrated model of multiple-condition ChIP-seq data reveals predeterminants of Cdx2 binding”, has been accepted for publication. The manuscript will be presented at the RECOMB 2014 conference, which will be held in Pittsburgh this year from April 2nd – 5th. As part of the conference submission, the manuscript was also reviewed in parallel by PLoS Computational Biology, and has now been accepted for publication there.
Our group manuscript, entitled “An integrated model of multiple-condition ChIP-seq data reveals predeterminants of Cdx2 binding”, has been accepted for publication. The manuscript will be presented at the RECOMB 2014 conference, which will be held in Pittsburgh this year from April 2nd – 5th. As part of the conference submission, the manuscript was also reviewed in parallel by PLoS Computational Biology, and has now been accepted for publication there.
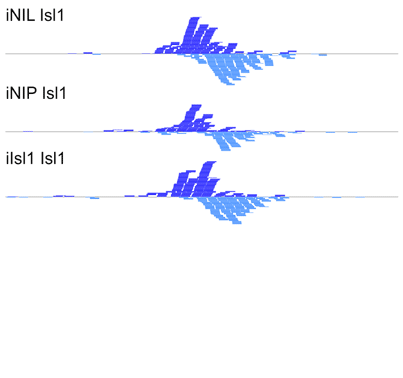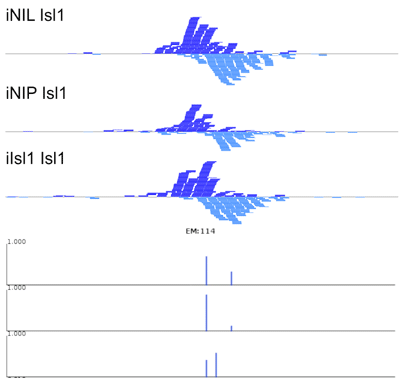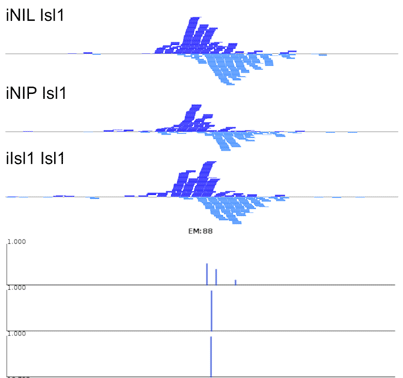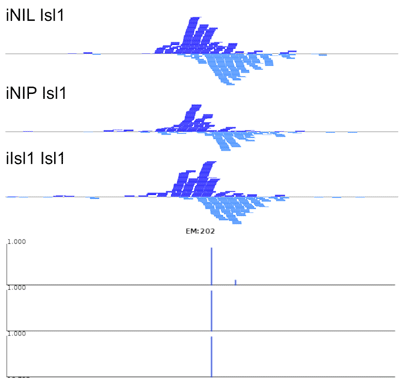
The manuscript describes MultiGPS, our new integrated machine learning approach for the analysis of multiple related ChIP-seq experiments. MultiGPS is based on a generalized Expectation Maximization framework that shares information across multiple experiments for binding event discovery, and enables the simultaneous modeling of sparse condition-specific binding changes, sequence dependence, and replicate-specific noise sources. MultiGPS encourages consistency in reported binding event locations across multiple-condition ChIP-seq datasets and provides accurate estimation of ChIP enrichment levels at each event.
The manuscript also demonstrates the advantages of MultiGPS with an analysis of Cdx2 binding in three distinct developmental contexts. By accurately characterizing condition-specific Cdx2 binding, MultiGPS enables novel insight into the mechanistic basis of Cdx2 site selectivity. Specifically, the condition-specific Cdx2 sites characterized by MultiGPS are highly associated with pre-existing genomic context, suggesting that such sites are pre-determined by cell-specific regulatory architecture. However, MultiGPS-defined condition-independent sites are not predicted by pre-existing regulatory signals, suggesting that Cdx2 can bind to a subset of locations regardless of genomic environment.
This work is a collaborative effort across multiple groups. MultiGPS was designed in a close collaboration between Shaun and joint-first author Matt Edwards (graduate student in David Gifford’s lab at MIT). Cdx2 ChIP-seq data in multiple cell types was generated by Esteban Mazzoni, Rich Sherwood, and Hynek Wichterle‘s lab. Akshay Kakumanu in our lab provided a critical evaluation of the performance of our methodologies, and was integral in getting the results wrapped up in time for RECOMB submission.
MultiGPS is open source software, available here.